 Please note: Given the multiple versions of TORTOISE with slight updates, TORTOISE_V3* will be used to denote different versions in this documentation as overall instructions/steps to run each command from different modules have not changed between versions. If you are starting a new study, please use the latest version of TORTOISE released.
Please note: Given the multiple versions of TORTOISE with slight updates, TORTOISE_V3* will be used to denote different versions in this documentation as overall instructions/steps to run each command from different modules have not changed between versions. If you are starting a new study, please use the latest version of TORTOISE released.
Some helpful tips before importing data:
- Please make sure that the folder to be imported contains only diffusion data. If you are not sure, please convert the dicoms from the raw folder to nifti using the mricron dcm2nii tool or if AFNI is installed then dcm2niix_afni. This should convert diffusion folder into nii with associated bvals and bvecs. If structural was present in the same folder, it would not have an associated bvals and bvecs file. This exercise is primarily for cases where diffusion data and other structural data are saved in the same raw folder and not distinguishable by data type.
- Before import, especially for a new study or studies with frequent scanner software updates, it is advised to open the raw diffusion dicom or .nii file in available visualization software. This is to ensure that the raw data has expected number of volumes as per recommended protocol.
- Once you import the raw data, please make sure you check your data before processing, mainly to ensure gradient information has been correctly translated from header/bvals-bvecs file. The reliable way to check the gradient information is to check the glyph maps or the non symmetric heurestic color map (tool included), detailed in "section 5- check your import results". The import tool relies on information present in the header file and if it is incorrect it will be migrated through the data and subsequent processing. So the initial checking, especially for new studies and studies with frequent scanner upgrades, is highly recommended.
- The new import tools is capable of reading direction information from the header. Although, if your data contains intermediate b values or custom sequence then please provide a gradient text file that is scaled to the maximum bvalue. Occasionally, while the data may have been acquired with the custom sequence, the header may have information stored with default settings. If you notice a discrepancy that may have resulted in incorrect import or bvals/bvecs output for your study, please provide the gradient text file during import. Providing the gradient file in such scenarios can ensure that the correct information is used for import and not rely on the information stored in the dicom header. The gradient text file will have tab separated three columns, with the same number of rows as the total volumes in the data to be imported. The top row will have the number of rows in the text file listed.
3.1. TORTOISE Version 3.1.3 and above versions- Import
All the executables can be accessed from the tortoise version bin folder: ./TORTOISE_V3*/DIFFPREPV3*/bin/bin folder if this has not been added to your path variable.
3.2. ImportDICOM
Type in the command line the following to see the following help file in the terminal ./ImportDICOM
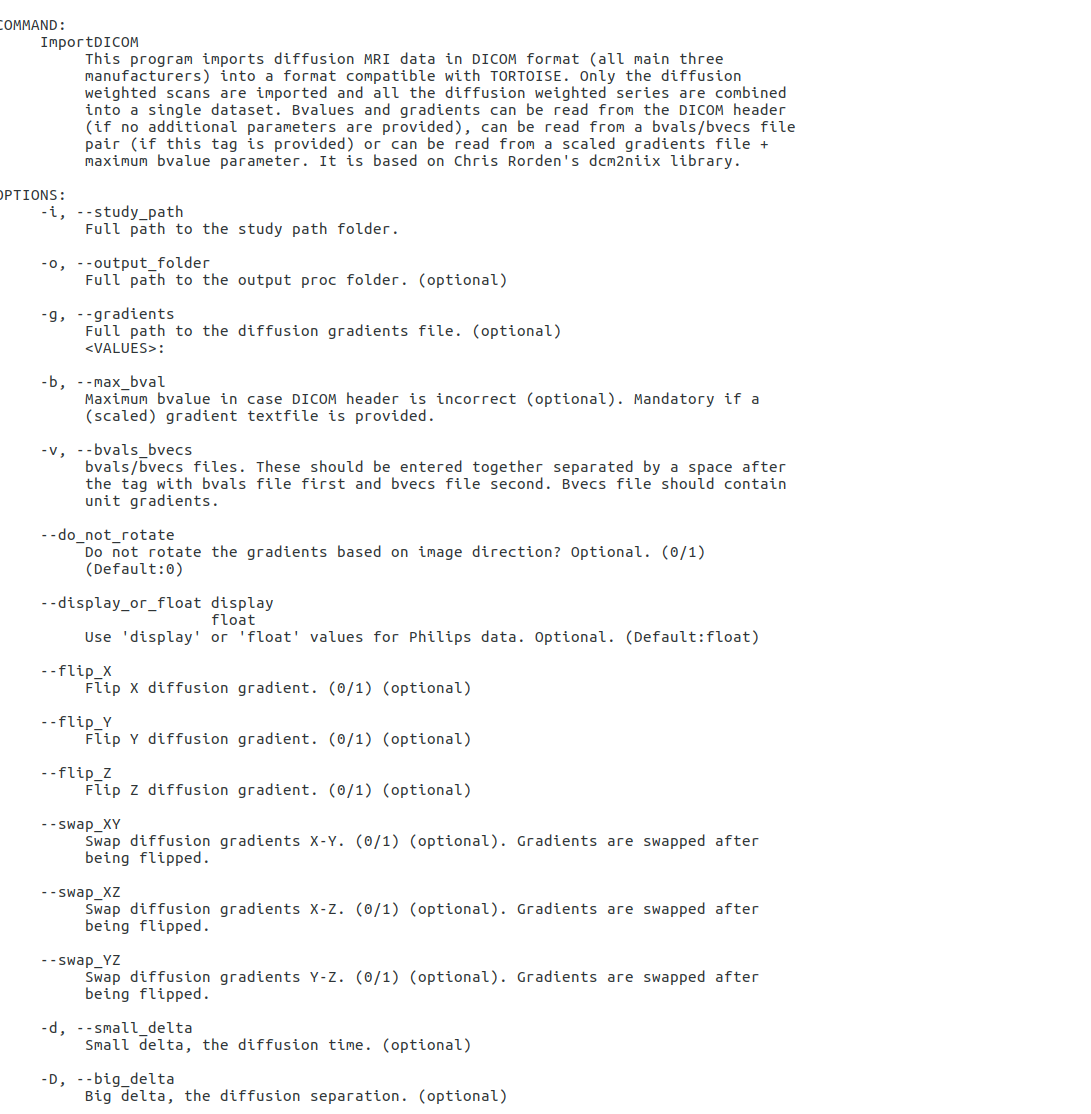
Generally, merely using ImportDICOM command without additional tags will suffice to import the data. However, in scenarios where you notice error in import after examining the data using tips detailed in section 5 of this documentation, additional tags detailed under the ImportDICOM help maybe used in import.
3.3. Import PARREC
Type in the command line the following to see the following help file in the terminal ./ImportPARREC, if your philips data has been saved in PAR REC format.
3.4. Import Bruker
Type in the command line the following to see the following help file in the terminal ./ImportBruker to show the help file to run this import.
