Check your import results using tools such as ComputeNSDECMap or ComputeGlyphMaps. The outputs from running the executables will be in the *_proc folder to be viewed using any visualization software that supports the viewing of RGB format files. You will find the executable in the ./TORTOISE_V3*/DIFFCALC/DIFFCALCV3*/ folder. To use either of the two commands, a tensor fitting needs to be performed. For the purpose of checking import results a linear fitting will suffice and the syntax is as follows:
-
Linear fitting should be performed to output a DT.nii
Usage: EstimateTensorWLLS full_path_to_listfile full_path_to_mask_file (optional) dti_bval_cutoff (optional. all volumes will be used)
After the computation of the DT.nii file, you can compute NSDEC map:
-
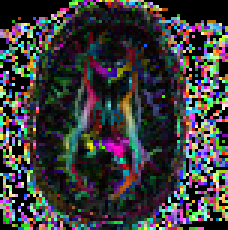
Non symmetric heuristic maps You can check the non symm heuristic map to determine if the data has been imported correctly by checking the non symm directionally encoded color maps.
Usage: ComputeNSDECMap full_path_to_tensor_image
GLYPHS
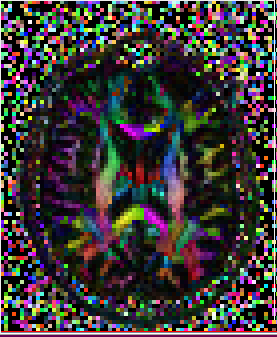
Tensor glyphs can also be viewed to determine if the import is correct
Usage: ComputeGlyphMaps full_path_to_tensor_NIFTI_file axis (optional) slice_number (optional) full_path_to_output (optional)
Axis: axial, sagittal or coronal. Default axial. slice_number: default center slice
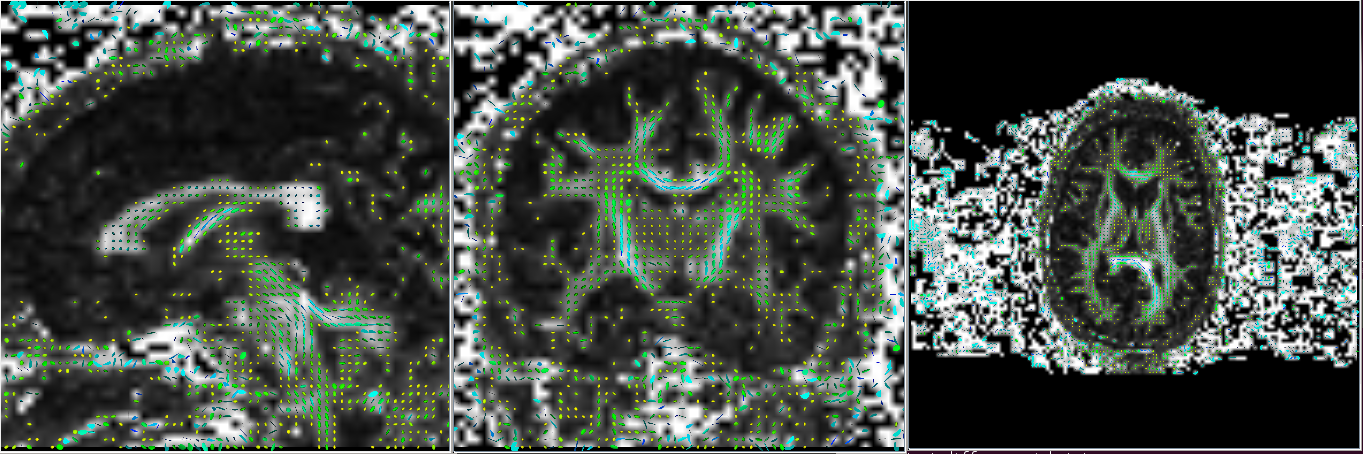
Gyphs can be useful in detecting if the data has been imported correctly. It is sometimes not sufficient to look at just the axial slice as the difference maybe more evident in the coronal and/or sagittal slices. Notice in the example below, the glyphs in the axial slice appears correct in both imports but the difference is seen in the coronal and sagittal slices.
Correct
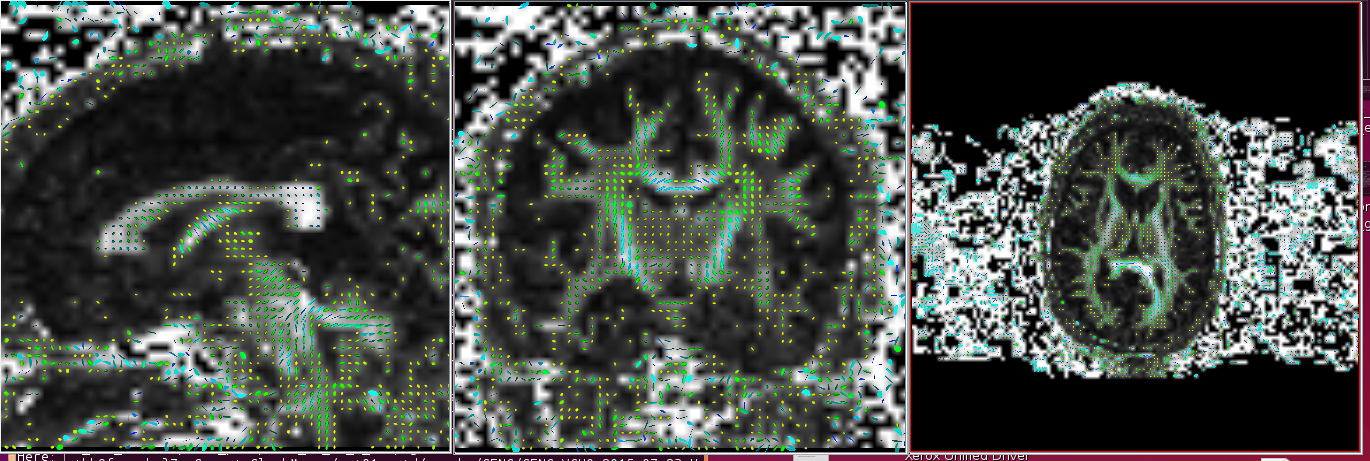
Incorrect (example when flip x and flip y are flipped)
Bmatrix check
It is essential to check the bmatrix file of the imported data. If it is a new study or if there has been a software upgrade at the scanner, it may affect the header information of the data. During such instances, it is advisable to ensure there has not been any change in the interpretation of the gradient file provided in the acquisition and how this information is stored in the header. The following quality check is advice to be performed periodically on a study, especially if it lasts many years.
- You may just convert the bmtxt file in the newly created _proc folder to bvals and bvecs to check if the information from the header has been imported correctly. You can use the command from the DIFFPREP/bin/bin folder TORTOISEBmatrixToFSLBVecs to convert the bmtxt file to bvals and bvecs. Please ensure the bvals and bvecs have values that are in line with the study protocol.
- Bmatrix check is crucial in blip up and blip down acquisition and to use DRBUDDI correction.