Hi, I used the Tortoise 3 to generate tensor and the eigenvector.
How can I visualize these images to check whether the final gradient table is corrected or not.
These images cannot be read appropriately by fsleyes or fslviews.
Thanks
Problem of the direction after DIFFPREP
Hi, Amritha,
Thanks for you reply.
By using your suggestions, I checked the results of DWI data after being processed by DIFFPREP.
However, I met a problem:
Before DIFFPREP, the gradient table is correct (checked with your method mentioned above as well as the eigenvector from FSL)
After DIFFPREP, the reoriented gradient table is NOT correct (seem not matched with the reoriented DWI images), both the DEC and eigenvector had wrong directions.
Here is the command I used:
DIFFPREP -i dwi.list --will_be_drbuddied 0 --do_QC 0 -d off --is_human_brain 0 -e ANTSSyN --high_b 0 --gibbs_ringing_correction off -s struct.nii -o dwi_DMC.list
Here is the data input (dwi.list and files):
https://pitt.box.com/s/buq5aq5cos68u405mds291n1vylpymo3
Here is the data output (dwi_DMC.list and files):
https://pitt.box.com/s/5sdurmtomwi6ks5sg83kuoezsnj8qhh1
Note that in the input, there is a structural image so that I can use DIFFPREP to reorient the DWI to the structural image. (it will be big reorientation)
I think the DIFFPREP did the correct reorientation on the DWI image, but may be incorrectly reorient bmtxt (gradient table).
I would be appreciated if you could take a look at the problem.
Thank you in advance!
Cirong
Re:Hi
Hi Cirong,
To better help guide the QC procedure, especially given this is animal data, it will help us to know the type of scanner that was used in the acquisition of this data. Was this data acquired on a Bruker scanner?
I have now processed the data and the results look similar to the results you have provided in the output folder. As you mentioned, there has been a large reorientation of DWI to match the structural orientation and it is registered well. What procedure did you use to bring the structural in what appears to be an ACPC or axialized orientation?
Can you please elaborate " After DIFFPREP, the reoriented gradient table is NOT correct (seem not matched with the reoriented DWI images), both the DEC and eigenvector had wrong directions." ?
Thanks,
Amritha
Hi,…
Hi, Amritha,
Thank you for your reply.
Yes, the data was acquired by Bruker with PV6.
The DWI image and the structural image registered well.
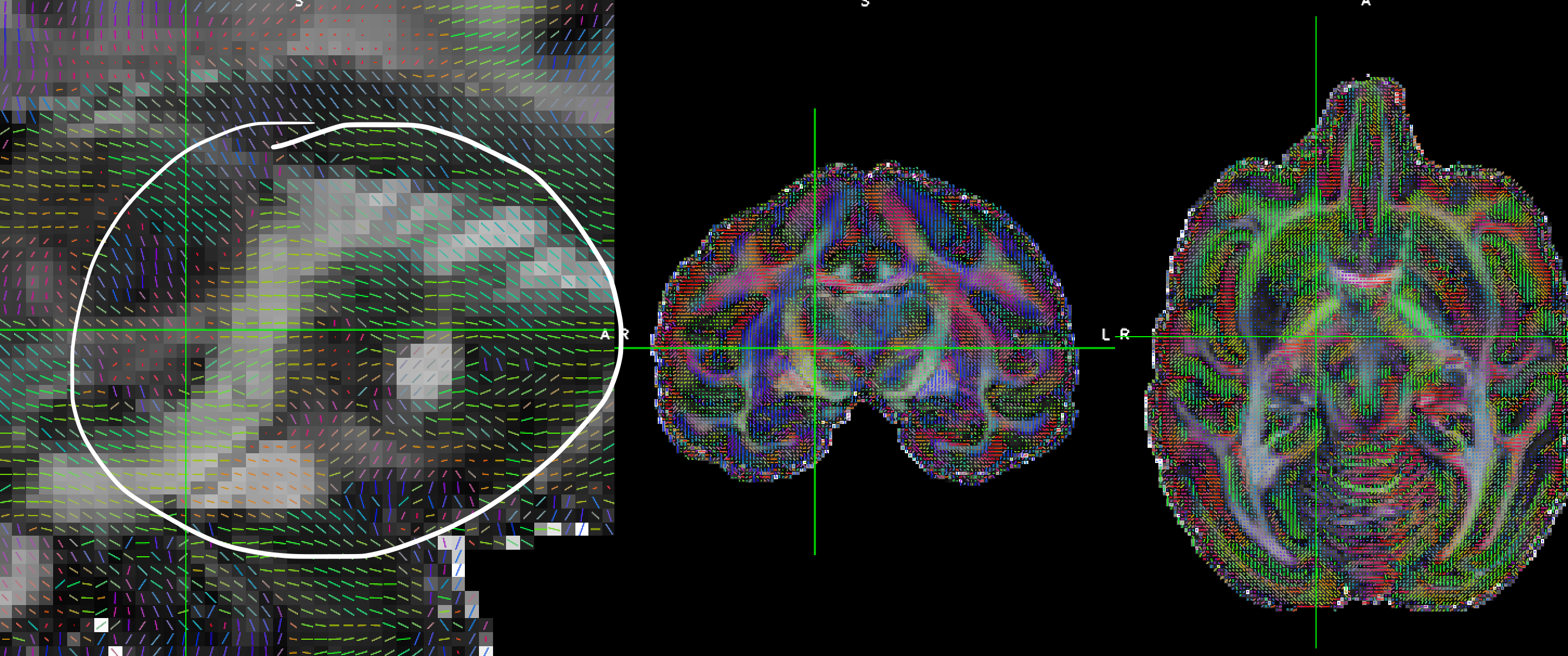
But if you used the reoriented gradient table to fit the reoriented DWI image, the resulting eigenvectors are wrong.
For example, in the screenshot, the direction should go up, not go down.
For the unprocessed data, the direction are corrected; and I used other software (mrtrix3) to do the registration, the resulting reoriented gradient table is also correct.
But the TORTOISE seems reoriented gradient table incorrectly. (at least not in the same way of reoriented the DWI image during the registration).
Hi
Sorry for the late reply.
Things have been crazy lately so I just had a chance to look at your data.
Unfortunately the import is wrong. So my first question is how did you import the data? Was it with ImportBruker executable?
The x and y gradients needed to be flipped. After that the processing was fine.
I am attaching the bmatrix file I used as an xlsx file. The system did not allow me to attach the dwi.bmtxt file directly.
So copy the contents of this file into dwi.bmtxt and reprocess your data.
Let us know.
Hi, Okan, Thank you very…
Hi, Okan,
Thank you very much!
I will have the matrix tested.
Cirong
Hi, Okan, I think there…
Hi, Okan,
I think there may be a potential bug/issues in TORTOISEBmatrixToFSLBVecs (converting bmtxt to fsl-grad) and maybe also in ImportNIFTI (converting fsl grad to bmtxt).
For example, if I used TORTOISEBmatrixToFSLBVecs to convert the grad file "you provided" and fitted the dwi data using "FSL", the resulting tensor has wrong orientations. (You can test it with your corrected bmtxt and fit the data using FSL)
On the contrary, if I used the original bvecs "I had" to fit the dwi data using "FSL", the resulting tensor is corrected (see my uploaded files previously); but this original bvecs resulted in the problem I mentioned previously.
Thanks!

Hi,
The recommendation would be to use the latest version of TORTOISE i.e ver 3.2.0, that has all the functionalities of DIFFCALC in command line.
To QC*, from DIFFCALCV320, you can generate absolute directionally encoded color (DEC) map using ComputeDECMap, nonsymmetric directionally encoded color (NSDEC) map using ComputeNSDECMap. In addition, you can generate glyph maps using the command ComputeGlyphMaps**.
DEC map generated would be the conventional color map.
Please refer to the following page of the documentation to guide you through interpreting NSDEC map and Glyph maps.
https://tortoise.nibib.nih.gov/tortoise/v313/5-check-your-import-results
In the documentation, input to these commands is the diffusion tensor (DT) of unprocessed data . As per your question, the input would be the DT of the processed data.
The glyph maps would be generated as png files. The DEC and NSDEC map would be in nifti format. You may visualize them in ITKSNAP, AFNI, MIPAV, to name a few that support the display of RGB format.
*Please note: Such QC are recommended to be done after import, before processing, as well, to ensure the associated gradient table/bvecs/bvals, the data have been imported correctly.
* *please refer to the following thread on the community page if you have trouble executing the ComputeGlyphMaps command : https://tortoise.nibib.nih.gov/community/core-dump-computeglyphmaps
Hope this helps,
Amritha