TORTOISE
TOLERABLY OBSESSIVE REGISTRATION AND TENSOR OPTIMIZATION INDOLENT SOFTWARE ENSEMBLE
TORTOISE has been migrated to TORTOISEV4 with BIG changes to both the software and the Website.
Please refer to the GitHub page for Download Instructions: https://github.com/QMICodeBase/TORTOISEV4
Please refer to the AFNI forums for the TORTOISE message board: https://discuss.afni.nimh.nih.gov/c/tortoise-message-board/9
This page will remain alive for Legacy reasons for users still using TORTOISE V3 or earlier. New users are encouraged to use TORTOISEV4.
The information below is ONLY for TORTOISEV3 and does not apply to the new TORTOISEV4.
What is TORTOISE?
The TORTOISE software package is used for processing diffusion MRI data. It contains four main modules:
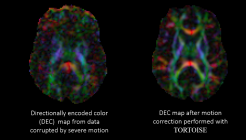
DIFF PREP is used for image resampling, motion, eddy current distortion, and EPI distortion correction using a structural image as target, and for rigid body re-orientation of single subject data to a common space.
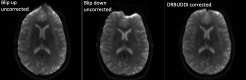
DR BUDDI is used for EPI distortion correction using pairs of diffusion data sets acquired with opposite phase encoding (blip-up blip-down acquisitions).
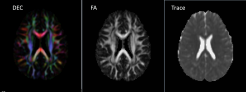
DIFF CALC software is used for tensor fitting, error analysis, directionally encoded color map visualization and ROI analysis
DR TAMAS (Diffeomorphic Registration for Tensor Accurate alignMent of Anatomical Structures) is a novel framework for inter-subject registration and template creation from Diffusion Tensor Imaging (DTI) data sets.
TORTOISEV4 IS COMING!!!!!!!!!
Check out https://github.com/QMICodeBase/TORTOISEV4 for early access.
How To Cite
TORTOISE Methods Publications
Please cite the following work if you use TORTOISE
- C. Pierpaoli, L. Walker, M. O. Irfanoglu, A. Barnett, P. Basser, L-C. Chang, C. Koay, S. Pajevic, G. Rohde, J. Sarlls, and M. Wu, 2010, TORTOISE: an integrated software package for processing of diffusion MRI data, ISMRM 18th annual meeting, Stockholm, Sweden, abstract #1597, (pdf)
- Mustafa Okan Irfanoglu1,2,3, Amritha Nayak1,2,3, Jeffrey Jenkins1,2, and Carlo Pierpaoli1,2 ,TORTOISEv3:Improvements and New Features of the NIH Diffusion MRI Processing Pipeline, ISMRM 25th annual meeting, Honolulu, HI, abstract #3540,(pdf)
In addition to the above two citations, if you use DRBUDDI, please cite the following:
- Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C.DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions, Neuroimage. 2015 Feb 1;106:284-99. doi: 10.1016/j.neuroimage.2014.11.042. Epub 2014 Nov 26. (pdf)
In addition to the TORTOISE citations, if you use DRTAMAS, please cite the following:
- Irfanoglu MO, Nayak A, Jenkins J, Hutchinson EB, Sadeghi N, Thomas CP, Pierpaoli C, DR-TAMAS: Diffeomorphic Registration for Tensor Accurate Alignment of Anatomical Structures. Neuroimage. 2016 May 15;132:439-454. doi: 10.1016/j.neuroimage.2016.02.066. Epub 2016 Feb 28. (pdf)
Acknowledgements
EXTERNAL SOFTWARE SOURCES
We have made use of the following external software sources, and would like to thank their creators. Masking in TORTOISE uses the bet2 algorithm from the FMRIB group, available in FSL. The non-linear tensor fitting algorithm uses the mpfit routine from the Markwardt IDL library, written by Craig Markwardt. The ImportDICOM routine is based on dcm2niix from Chris Rorden.