Dear all experts,
When I used ImportDICOM to import diffusion dataset, the b vector in the generated bvecs file is not consistent with the b vector table I used. I have no idea about that and I do not know if it is related to the coordinate system (XYZ or "phase read slice") ImprotDICOM used.
I acquried diffusion dataset on Simens 3T prisma and the coordinated system I used in the b vector table is "prs" (like the following picture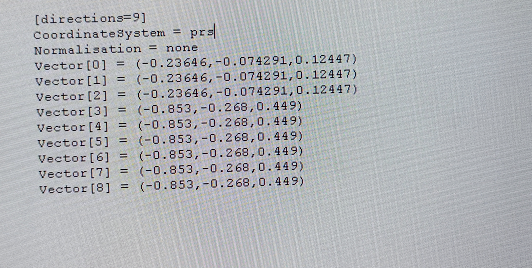 ).
).
The b vector I used is "[-0.853 -0.268 0.449]", but the read b vector used ImportDICOM is "[0.268 0.853 0.449]".
Looking forward to your reply and thanks very much.
Best,
Zhaoqing
Correct
The GlyphMaps seem correct. The fact that the coronal and sagittal dont have full coverage is weird. It is probably because of a bug and is not expected.
The interpretation of the NSDECMaps can be found in https://tortoise.nibib.nih.gov/tortoise/v313/5-check-your-import-results

Hello,
I assume you are only asking about the coordinate system of the diffusion gradients and not the image. (The image is in scanner physical coordinate system with a POSITIVE orientation determinant).
The coordinate system of the diffusion gradients is actually neither the scanner or FOV coordinate system.
IT is: g_x: from left of the image towards right
g_y: from bottom of the image towards top
g_z: from first slice towards last
In your case, your gradients are defined in phase-read-slice. I actually recommend xyz system for Siemens scanners but not a big deal.
Because in prs system (for AP and PA phase-encoding), the first gradient component is vertical and the second is horizontal, the import routine swaps these two gradients and checks for the negativity of the signs.
I strongly suggest to do a quick tensor fitting (with EstimateTensorWLLS) on the imported data and use the ComputeGlyphMaps and ComputeNSDECMap executables on the comptued tensors and make sure everything is okay as described on the website.
Hope this helps.