Hi guys,
I apologise if I'm posting a question that was already answered.
I'm facing a problem after I run DIFFPREP in one subject of a project I'm collaborating with. I had to create a T2 image from a T1 using fat_proc_imit2w_from_t1w which worked pretty fine. Then I ran the script provided by Taylor for the AFNI bootcamp, that worked fine for all other subjects except for this one.
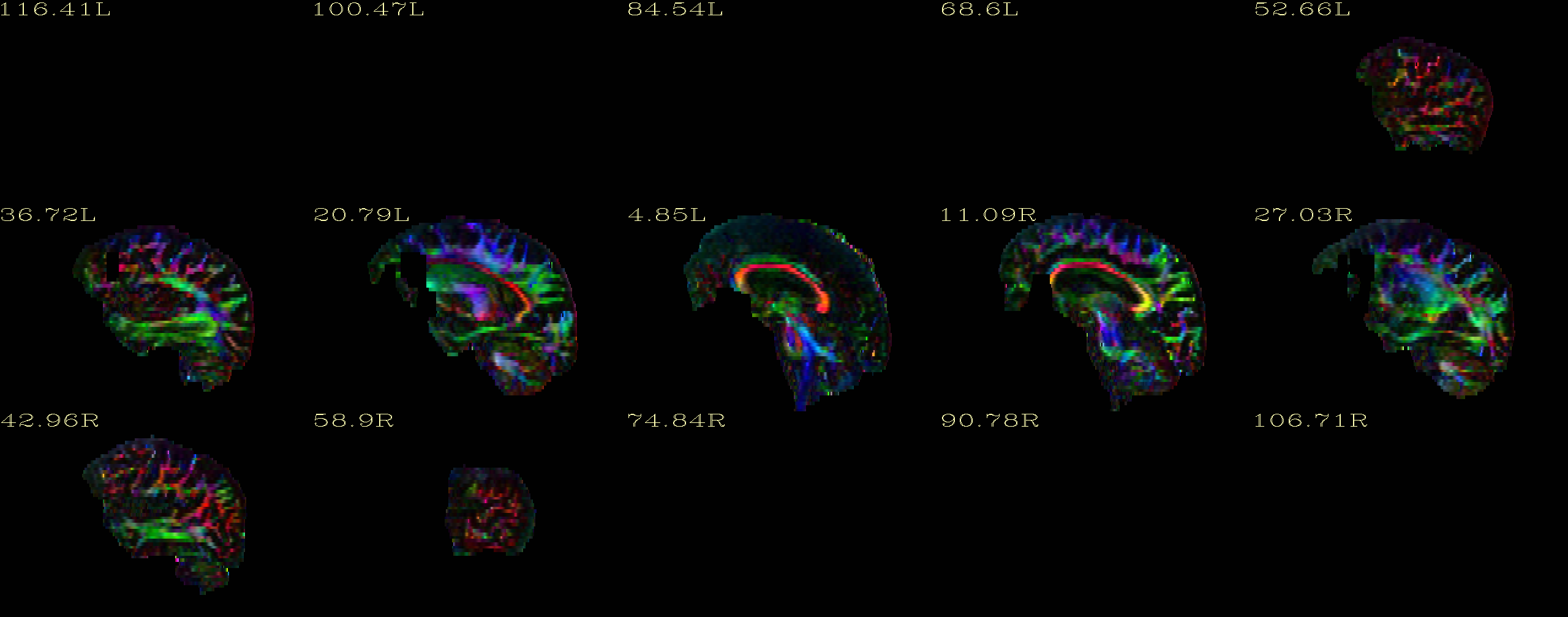
My DTI and T2 data look fine before running DIFFPREP. After I ran DIFFPREP, I checked the DEC maps for quality control and there are missing clusters in the frontal lobe (apparently, DIFFPREP erases them). I'm sending attached the DEC images I mentioned.
Could you please help me with a correction of this problem?
Thank you in advance,
Wyllians Borelli
Hello Amritha, I used the…
Hello Amritha,
I used the command fat_proc_decmap to create the DEC map, using the mask generated by fat_proc_dwi_to_dt.
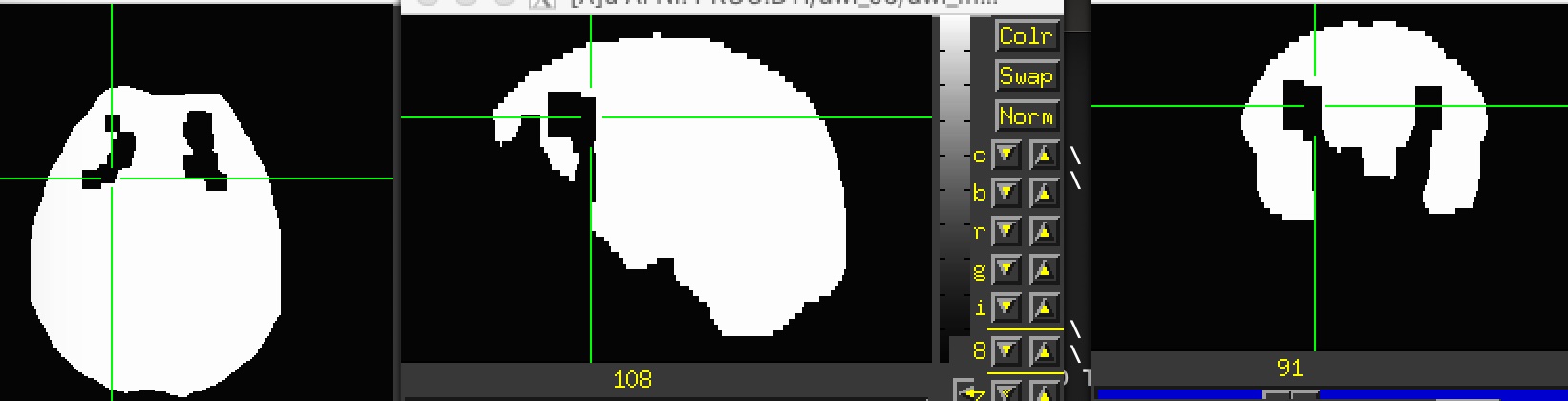
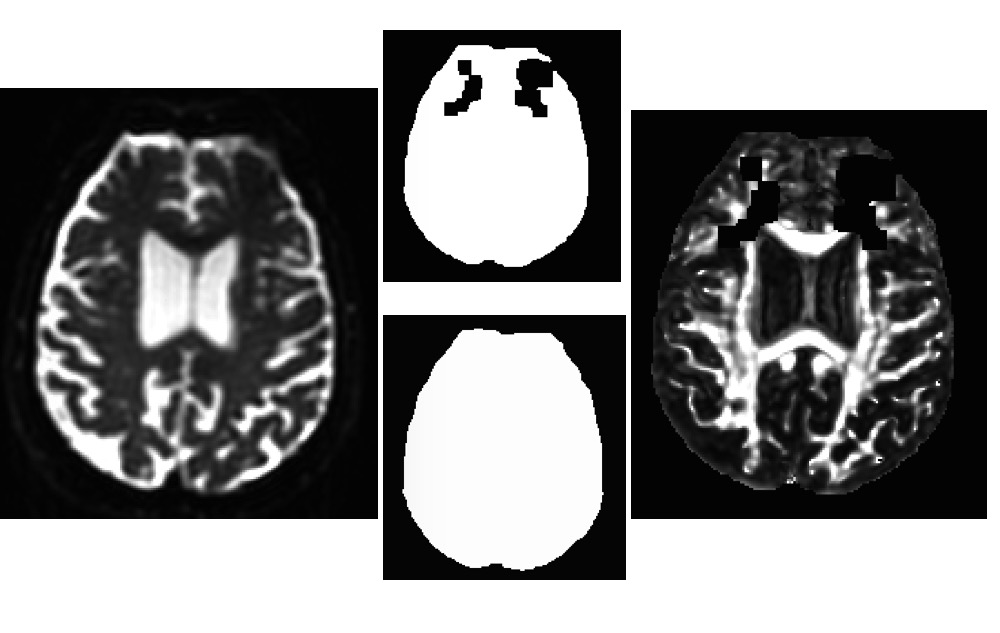
This mask has the same missing clusters (attached), so I tried to manually fill it (using AFNI's draw dataset) and rerun the fat_proc_decmap. Apparently, it didn't work. I also tried to provide this filled_mask in the fat_proc_dwi_to_dt but it failed again.
Am I doing something wrong in this process?
Thank you,
Wyllians
Hi, Willians- Can you…
Hi, Willians-
Can you please post your fat_proc_dwi_to_dt command, as well as your fat_proc_decmap command? My guess is that the issue is with the masking (as Ami noted).
fat_proc_dwi_to_dt will use 3dAutomask on the [0]th volume of the DWI set if no masking options are given; this can be problematic on DWI dsets, since they have have a huge amount of brightness inhomogeneity. Using "-mask_from_struc" is generally the recommended way to go, but it is possible this might not always get things right, again depending on the dataset's potential quirks (sigh).
--pt
fat_proc_dwi_to_dt
Hi Taylor,
Here's the command I used (based on the Tortoise class, AFNI bootcamp in September 2018):
# =============== STEP 10: tensor + param estimation ===============
# shortcut for tort path
set itort = $path_P_ss/dwi_03_ap
set odir = $path_P_ss/dwi_05# do autoflip check: not ideal to need this...
@GradFlipTest \
-in_dwi $itort/ap_proc.nii \
-in_col_matT $itort/ap_proc.bmtxt \
-prefix $itort/GradFlipTest_rec.txtset my_flip = `cat $itort/GradFlipTest_rec.txt`
fat_proc_dwi_to_dt \
-in_dwi $itort/ap_proc.nii \
-in_col_matT $itort/ap_proc.bmtxt \
# -in_struc_res $itort/structural.nii \
-in_ref_orig $path_P_ss/anat_01/t2w.nii \
-prefix $odir/dwi \
# -mask_from_struc \
$my_flip# =============== STEP 11: tensor DEC (color) maps =================
fat_proc_decmap \
-in_fa $odir/dt_FA.nii.gz \
-in_v1 $odir/dt_V1.nii.gz \
-mask $odir/dwi_mask.nii.gz \
-prefix $odir/DEC
I also tried to provide a mask using "-mask" but it didn't work. As I mentioned, I didn't use "-mask_from_struc" because I was not using FreeSurfer for this dataset. This command worked for all other subjects. If you suggest running FreeSurfer for this subject, then I'll do it. I've attached both masks I tried to use.
Best,
Wyll
Hi, Wyllians- Thanks for…
Hi, Wyllians-
Thanks for posting the commands. I am a bit confused about why you aren't using "-in_struc_res $itort/structural.nii" (and therefore also not "-mask_from_struc")? The file structural.nii should just be output by TORTOISE, and not related to running FreeSurfer.
Also, in what you post, there is one mask with gaps, and one mask without gaps-- what made the mask without gaps?
--pt
Hi Taylor, Actually I made…
Hi Taylor,
Actually I made a mistake. I thought that "-in_struc_res $itort/structural.nii" was output by FreeSurfer. I had a problem with my TORTOISE script (I used ap_proc instead of DMC_structural.nii).
That mask was filled using AFNI's GUI (draw dataset).
Problem solved. Thank you very much.

Hi Wyllians,
Did you provide any mask in the generation of your tensor file (DT) file? Which command did you use to generate the DEC map?
Amritha